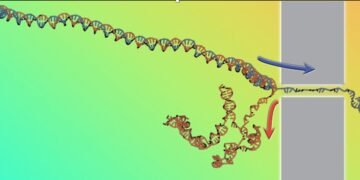
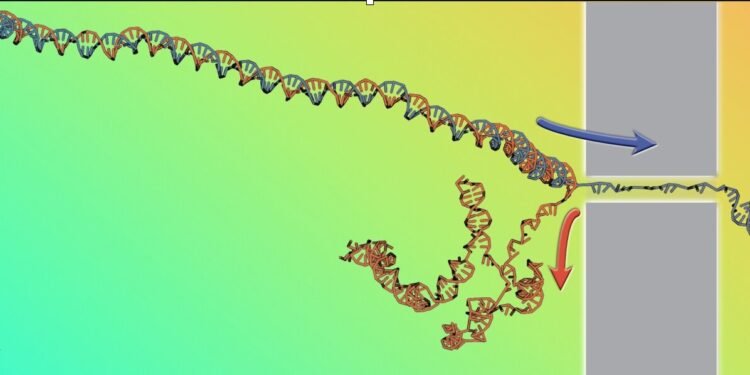
Understand how the parts of a molecule interact with each other knowing only how the molecule behaves when it is deformed or destroyed. That is the challenge of the research carried out by Cristian Micheletti of SISSA and recently published in the Physical Research Letters. Specifically, scientists studied the opening of the DNA double helix when it was pulled quickly through the nanopore, reconstructing the main thermodynamic properties of DNA from the speed of the process alone.
The insertion of polymers through nanopores has long been studied for complex applications and for its importance in many areas, including genome sequencing. The latter is based specifically on forcing the DNA to pass through a narrow gap so that only one of the two strands of the double helix can fit into it, while the other strand will be at the back. DNA inserted into a nanopore by an electric current will be forced open and exposed, an effect known as “decompression”.
The research team, which also includes lead author Antonio Suma from the University of Bari and Vincenzo Carnevale from Temple University, used a computer cluster to speed up the process in different ways while saving the amount of DNA decompression. this type of data is directly available in experiments but few studies so far.
Using previously developed theoretical and mathematical models, the researchers were able to perform a ‘reverse’ operation, using velocity information to retrieve the thermodynamics of the double helix formation and collapse.