A New Sensor Mimics the Function of Cell Membranes, the device identifies specific genes such as cell receptors and can lead to the diagnosis of cancer and other diseases in a systematic way.
Taking inspiration from the body’s sensory system, an MIT-led team has developed a new sensor that can detect the same molecules that natural receptors can recognize. In a project that combines several new technologies, researchers have developed a prototype sensor that can detect an inhibitor called CXCL12, at ten or hundreds of parts per billion.
This is the first important step in the development of a method that can be used to perform routine examinations for cancer that is difficult to detect cancer or metastatic tumors, or as a highly biomimetic electronic “nose”, according to the researchers.
“Our hope is to create a simple device that allows you to test at home, with high definition and understanding. The earlier you find the cancer, the better the treatment, so during the cancer research is an important part that we want to fight,” said Shuguang Zhang, head of research at MIT’s Media Lab.
The device is inspired by the membrane that surrounds all cells. Within these membranes are thousands of receptor proteins that detect molecules in the environment. The MIT team modified some of these proteins so that they could survive outside the membrane and placed them in a crystalline protein layer on top of a network of graphene transistors.
When a target molecule is detected in the sample, these transistors transmit the information to a computer or smartphone. This type of sensor can be used to monitor any body fluid, such as blood, tears or water, according to the researchers, and can monitor many different targets at the same time, depending on the type of proteins that respond to it.
Rui Qing, a former MIT researcher who is now a professor at Shanghai Jiao Tong University, said, “We are identifying receptors that are sensitive to biological systems and putting them in a bioelectronic interface, which allows us to bring all these signals and convert them into electrical outputs that can be analyzed and interpreted by machine learning algorithms.
Qing and Mantian Xue PhD ’23, are the lead authors of the study, which appears today in Science Advances. Along with Zhang, Tomás Palacios, director of MIT’s Microsystems Laboratory and professor of electrical engineering and computer science, and Uwe Sleytr, professor emeritus at the Institute for Synthetic Bioarchitectures at the University of Natural Resources and Life Sciences in Vienna, are the lead authors of the paper.
Without membranes
Most current research sensors rely on antibodies or aptamers (small strands of DNA or RNA) that can capture specific target cells from fluids such as blood. However, these two methods have limitations: the water can easily degrade the aptamers, and creating antibodies so that each batch is the same can be difficult. Another method that scientists have explored is the creation of sensors based on protein receptors in cell membranes, which cells use to monitor and react to their environment. The human genome codes for thousands of these receptors. However, the receptors are difficult to work with because once they are removed from the cell membrane, they retain their structure if stopped in the soap.
In 2018, Zhang, Qing and others reported a new method to convert hydrophobic proteins into water-soluble proteins, by replacing some hydrophobic amino acids with hydrophilic amino acids. This system is called the QTY code, when the letters represent the three hydrophilic amino acids – glutamine, threonine and tyrosine – which represent the hydrophobic amino acids leucine, isoleucine, valine and phenylalanine.
“People have been trying to use receptors for research for decades, but it’s difficult to use them on a large scale because receptors need soap to stabilize. The innovation of our method is that we can make water soluble and produce them on a cheap scale,” says Zhang.
Zhang and Sleytr, who have been working together for a long time, decided to work together to try to synthesize proteins that accept water nutrients on the surface, using bacterial proteins that Sleytr had studied for years. These proteins, known as S-layer proteins, are found in the upper layer of the cell envelope in many types of bacteria and archaea.
When S-layer proteins are crystallized, they form a monomolecular network that is interconnected at the surface. Sleytr has previously shown that these proteins can be combined with other proteins such as antibodies or enzymes. For this study, researchers, including scientist Andreas Breitwieser, who is also a co-author of the paper, used the S-layer protein to create a non-functional sheet of a type of receptor protein called CXCR4.
This receptor binds to a target molecule called CXCL12, which plays an important role in many human diseases, including cancer, and the HIV envelope glycoprotein, responsible for viral entry into human cells. “We use these S-layer systems that allow these active molecules to be attached to the surface in a monomolecular network, with a better distribution and organization,” says Sleytr. “It’s like a chess board where you can lay out the different pieces very well.”
The researchers called their sensing technology RESENSA (Receptor S-layer Electric Nano Sensing Array).
Effects on biomimicry
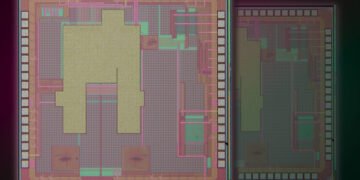
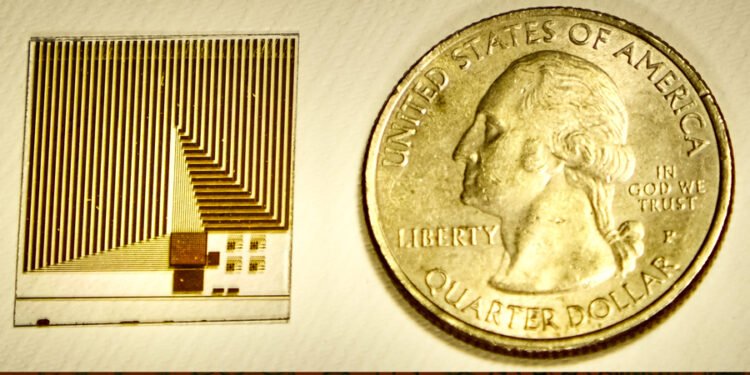
These crystal S-beds can be placed on almost any surface. For this application, the researchers combined the S-layer and chip with a graphene-based transistor system previously developed by Palacios’ lab. The unique atomic thickness of graphene transistors makes them ideal for the development of highly sensitive detectors.
Working in the Palacios lab, Xue adapted the chip so that it could be coated with a double layer of proteins – crystallized S-layer proteins attached to water-soluble receptor proteins. When a target molecule in the sample binds to a receptor protein, the charge on the target changes the electrical properties of the graphene in a way that can be easily interpreted and transmitted to a computer or smartphone connected to the chip.
“We chose graphene as the transducer material because it has good electrical conductivity, which means it can convert these signals very well. It has a very high-to-low ratio because it is a sheet of carbon atoms, so any changes on the surface, made by protein binding material, translate directly to the whole thing,” Xue explains.
A graphene transistor chip with a density of 1 trillion acceptors per square centimeter can be used with S-layer receptor protein to cover the surface.
This allows the explosion to obtain the maximum sensitivity that the receptors provide, within the clinical limits necessary for research in the human body. The chip integrates more than 200 devices, providing redundancy in signal detection that helps ensure reliable measurements even for particles, such as those that can reveal the presence of early brain damage or the onset of Alzheimer’s disease, the researchers said.
By using the QTY code, it will be possible to modify existing receptor proteins that can be used, the researchers believe, to create multiple sensors on a single chip to look at almost any molecule that cells can detect. “Our goal is to create the basic technology to enable wearable devices in the future to be able to connect to phones and computers, so you can test at home and quickly know if you need to go to the doctor,” Qing says.
“This new method is a combination of different research areas such as chemistry and materials, physics and electrical engineering, which makes the team’s process more efficient,” explains Piero Baglioni, professor of physical chemistry at the University of Florence, who was not involved in the study. “Also, I think this is an advance that could be very useful in diagnosing many diseases.”
Source: Massachusetts Institute of Technology