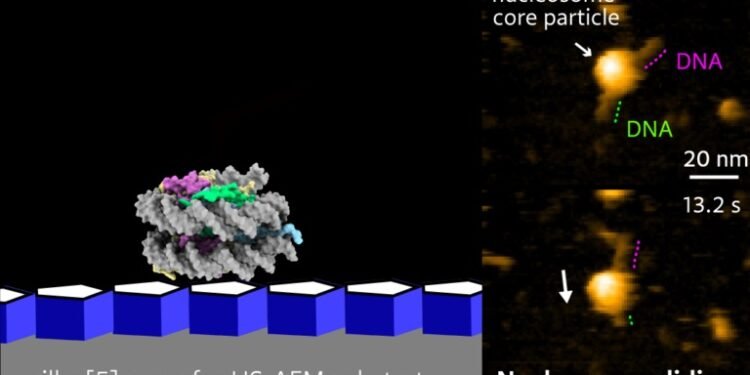
Researchers at Kanazawa University report in the Nano Letters that the discovery of a powerful biomolecular system can benefit genes. The structure, which was revealed (Nanoscale Biomolecular Shift) using high-energy atomic microscopy, includes DNA and carrier molecules.
In organisms whose cells have a nucleus, such as plants and animals, the main packaging material for DNA is nucleosomes. A nucleosome is made up of a piece of DNA wrapped around eight proteins called histones. Gene expression, which is the basis for protein production, requires “reading” DNA, in which DNA must be removed temporarily.
A comprehensive study, in detail, of DNA-histone and nucleosome dynamics is essential to better understand DNA unwinding and related processes. Mikihiro Shibata of Kanazawa University and his colleagues have now succeeded in video recording the nucleosome dynamics of H2A.Z, a different histone element associated with different biological processes.
The videos reveal the sudden release of H2A.Z nucleosomes. Different forms of the stone, such as H2AZ, differ from the canonical form (such as H2A) in stable nucleosomal packing. They form unstable nucleosomes with some biological activity; H2A.Z is thought to play a role in early embryonic development and stem cell differentiation. The dynamics of the H2A.Z nucleosome under physiological conditions is unknown. Shibata and colleagues use high-speed atomic force microscopy (HS-AFM) to study the dynamics of H2A.Z nucleosomes, as the method is a powerful nanoimaging tool for visualizing molecular structures and their dynamics in high space decision – time.
To observe DNA-histone dynamics in HS-AFM experiments, the nucleosome must be attached to the substrate. DNA should be easily transferred to the substrate, but at the same time, the DNA interaction is still weak so as not to interfere with the dynamic process. Therefore, scientists prepare natural materials by placing pillars [5] on mica. Pillar arenes[5], molecules with a pentagonal tubular structure, form thin films on mica and provide an ideal surface for monitoring nucleosome dynamics.
The researchers looked at the evolution over time of the composition of nucleosomes attached to DNA strands. Analysis of canonical H2A histones confirmed the stability of H2A nucleosomes: no significant changes over time. Analysis of histone H2A.Z variants, however, showed a different picture. HS-AFM with a time resolution of 0.3 s revealed an unwinding event, in which the nucleosome material falls along the DNA strand.
The findings of Shibata and his colleagues may lead to a better understanding of the biochemical mechanisms behind gene expression. According to the researchers: “The single-molecule HS-AFM imaging presented here may help clarify the relationship between nucleosome dynamics and gene structure… in the future close.”
Background
Advanced Atomic Force Microscope
The general principle of atomic force microscopy (AFM) is to probe the surface of a sample using a small tip. During this vertical (xy) analysis, the tip, which is attached to a small cantilever, follows the vertical profile (z) of the sample, making the force on the cantilever possible to measure. The degree of force in the xy position can be related to the z value; The xyz data generated during the survey are then translated into a high-resolution map that provides structural information about the sample being studied. In high-speed AFM (HS-AFM), the principle of operation is slightly more complicated: the cantilever is made to rotate in its frequency direction. As the tip is moved around the surface, changes in the amplitude (or frequency) of the cantilever oscillations – resulting from the interaction of the tip with the surface of the sample- are recorded, as they give a measurement of local “z” value. AFM does not include a lens, so its resolution is not limited by the so-called diffraction limit as in X-ray diffraction, for example.
HS-AFM produces video, where the time interval between frames depends on how fast one frame can be produced (per xy analysis of the sample). Researchers at Kanazawa University have developed the HS-AFM in recent years, so that it can be used to study biochemical and biomolecular processes directly. Mikihiro Shibata and his colleagues have now applied the method to study the dynamics of nucleosomes, revealing the mechanism of movement of nucleosomes along the DNA strand.
Source: Kanazawa University.